This post has two main purposes, 1) serves as personal notes for handling continuous features in decision tree; 2) try to use trait to add more computation operations to vectors, because the original Vec shipped with Rust is nowhere near the numpy in Python when it comes to scientific computation. (Though I know that Vec may not be designed to do handle such task.)
There are many ways to handle continuous descriptive features, such as discretions. This post will exploit weighted variance as a measurement for splitting continuous feature at a node.
The weighted variance is computed by the following equation, where $n$ is the number of rows in the data, $\mathcal{L}$ is the set of all unique labels, $D$ is the column with continuous features ($n$ rows), $p^*$ denotes the best split position in $D$.
$p^*=argmin_{p\in [1, n)} \sum_{l\in \mathcal{L}}\frac{\|\mathcal{L}_{i=l}\|}{\|\mathcal{L}\|} [var(D_{[0, p)}, \mathcal{L}_{i=l}) + var(D_{[p, n)}, \mathcal{L}_{i=l})]$
Once the algorithm decides the best split position of $D$, we can apply divide and conquer! For example, if $p^*$ has been computed, then we recursively apply this mechanism to both $D[0 .. p^*]$ and $D[p^* ..]$. When the split position arrays, $S_i$ and $S_j$, of $D[0 .. p^*]$ and $D[p^* ..]$ return, $S_i$ and $S_j$ will be merged and sorted as final value.
Let's try this algorithm on this small dataset,
| Continuous Data | Label |
| 0 | 0 |
| 2 | 0 |
| 3 | 1 |
| 7 | 0 |
| 15 | 0 |
| 35 | 2 |
| 45 | 1 |
| 47 | 1 |
| 55 | 0 |
| 57 | 1 |
| 61 | 2 |
| 67 | 2 |
| 81 | 1 |
| 92 | 0 |
| 96 | 2 |
| 97 | 2 |
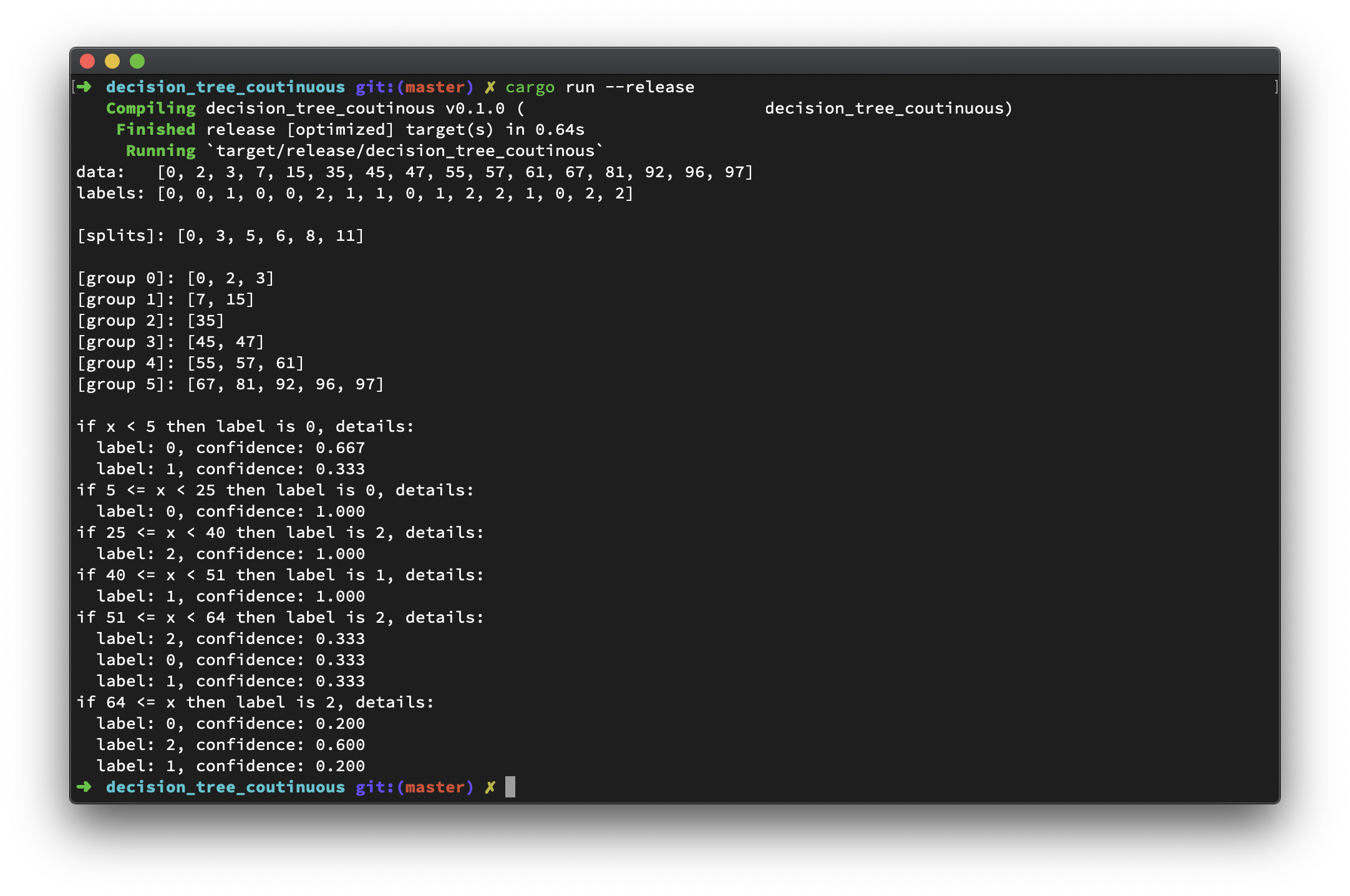
Looks good to me! And code goes below.
use std::collections::HashMap; extern crate num_traits; use num_traits::Num; struct Outcome { label: u32, left_boundary: Option<f64>, right_boundary: Option<f64>, detail: Vec<OutcomeDetail> } struct OutcomeDetail { label: u32, confidence: f64 } fn main() { // the final label of data let labels: Vec<u32> = vec![0,0,1,0,0,2,1,1,0,1,2,2,1,0,2,2]; // column with continuous data type let continuous_column: Vec<u32> = vec![0,2,3,7,15,35,45,47,55,57,61,67,81,92,96,97]; println!("data: {:?}", continuous_column); println!("labels: {:?}\n", labels); assert_eq!(labels.len(), continuous_column.len()); let count = labels.len(); // build decision tree with continuous column let splits = decision_tree_continuous(&continuous_column, &labels, 0, count); let outcome = compute_outcome(&continuous_column, &labels, &splits); // describe decision tree for current data println!("[splits]: {:?}\n", splits); let mut group = 0; for s in 0..splits.len() - 1 { println!("[group {}]: {:?}", group, continuous_column[splits[s]..splits[s + 1]].to_vec()); group += 1; } println!("[group {}]: {:?}\n", group, continuous_column[splits[splits.len() - 1]..].to_vec()); // describe decision tree for out in outcome { print!("if "); if let Some(left_boundary) = out.left_boundary { print!("{} <= ", left_boundary); } print!("x"); if let Some(right_boundary) = out.right_boundary { print!(" < {}", right_boundary); } println!(" then label is {}, details:", out.label); for d in out.detail { println!(" label: {}, confidence: {:.3}", d.label, d.confidence); } } } /// Build decision tree with continuous column /// /// @param column, column with continuous data type /// @param labels, the final label of data /// @param from, beginning position (included) /// @param to, ending position (not included) /// /// @return array of position that splits the data fn decision_tree_continuous(column: &Vec<u32>, labels: &Vec<u32>, from: usize, to: usize) -> Vec<usize> { if to - from <= 1 { // it is the only row left vec![to - 1] } else { // split rows in range [from, to) let split_pos = split_continuous_column(column, labels, from, to); if split_pos == from || split_pos == to { // pure leaf vec![split_pos] } else { // split the first part, range [from, split_pos) let mut sub_split_i = decision_tree_continuous(column, labels, from, split_pos); // split the second part, range [split_pos, to) let mut sub_split_j = decision_tree_continuous(column, labels, split_pos, to); // merge split positions sub_split_i.append(&mut sub_split_j); sub_split_i.sort(); sub_split_i.dedup(); sub_split_i } } } /// Split the column with continuous data type /// /// @param column, column with continuous data type /// @param labels, the final label of data /// @param from, beginning position (included) /// @param to, ending position (not included) /// /// @return split position fn split_continuous_column(column: &Vec<u32>, labels: &Vec<u32>, from: usize, to: usize) -> usize { let mut weighted_variances: Vec<f64> = Vec::new(); // try every single possible split position for i in from + 1..to { // the first part let column_i = column[from..i].to_vec(); // the second part let column_j = column[i..to].to_vec(); // get corresponding labels and occurrence of position in column_i and column_j respectively let (column_i_labels, occurrence_i) = get_unique_labels(&labels[from..i]); let (column_j_labels, occurrence_j) = get_unique_labels(&labels[i..to]); // weighted variance will be computed and used as metrics for the split let weighted_variance_i = weighted_variance(&column_i, &column_i_labels, &occurrence_i); let weighted_variance_j = weighted_variance(&column_j, &column_j_labels, &occurrence_j); let weighted_total_variance = weighted_variance_i + weighted_variance_j; weighted_variances.push(weighted_total_variance); } // the split with minimum weighted variance is preferred weighted_variances.argmin().unwrap() + from } /// Get unique labels and its location of presence /// /// # Example /// /// ```rust /// let labels = vec![0,1,0,1,0,1,2]; /// let (unique_labels, occurrence) = get_unique_labels(&labels); /// unique_labels.sort(); /// assert_eq!(unique_labels, vec![0,1,2]); /// /// assert_eq!(occurrence[0], vec![0,2,4]); /// assert_eq!(occurrence[1], vec![1,3,5]); /// assert_eq!(occurrence[2], vec![6]); /// ``` fn get_unique_labels(labels: &[u32]) -> (Vec<u32>, Vec<Vec<usize>>) { let mut unique: HashMap<u32, Vec<usize>> = HashMap::new(); for index in 0..labels.len() { if !unique.contains_key(&labels[index]) { let mut occurrence = Vec::new(); occurrence.push(index); unique.insert(labels[index], occurrence); } else { unique.get_mut(&labels[index]).unwrap().push(index); } } let mut unique_labels: Vec<u32> = Vec::new(); let mut label_occurrence: Vec<Vec<usize>> = Vec::new(); for (k, v) in unique { unique_labels.push(k); label_occurrence.push(v); } (unique_labels, label_occurrence) } /// Compute weighted variance /// /// @param column, column with continuous data type /// @param unique_labels, unique labels of the data in the given column /// @param corresponding_index, the location of presence of the unique labels /// @return weighted variance fn weighted_variance(column: &Vec<u32>, unique_labels: &Vec<u32>, corresponding_index: &Vec<Vec<usize>>) -> f64 { let unique_labels_count = unique_labels.len() as f64; let mut weighted_var = 0f64; for i in 0..unique_labels.len() { let sample_variance = column.select(&corresponding_index[i]).var(); weighted_var += corresponding_index[i].len() as f64 / unique_labels_count * sample_variance; } weighted_var } /// Compute outcome data /// /// @param column, column with continuous data type /// @param labels, the final label of data /// @param splits, splits of the continuous column fn compute_outcome(column: &Vec<u32>, labels: &Vec<u32>, splits: &Vec<usize>) -> Vec<Outcome> { let mut outcome: Vec<Outcome> = Vec::new(); let mut left_boundary = 0f64; let mut has_left_boundary = false; let mut splits = splits.clone(); splits.push(column.len() - 1); for s in 0..splits.len() { let mut current_outcome = Outcome { label: 0, left_boundary: None, right_boundary: None, detail: vec![] }; if splits[s] == 0 { continue; } else { if has_left_boundary { current_outcome.left_boundary = Some(left_boundary); } let current_split; let current_labels: &[u32]; if s == splits.len() - 1 { current_labels = &labels[splits[s - 1]..]; current_split = (column[splits[s - 1]] + column[splits[s - 1] - 1]) as f64 / 2.0; } else { current_labels = &labels[splits[s - 1]..splits[s]]; current_split = (column[splits[s] - 1] + column[splits[s]]) as f64 / 2.0; current_outcome.right_boundary = Some(current_split); } let (unique_label, occurrence) = get_unique_labels(current_labels); let sum = current_labels.len() as f64; let mut most_confidence = 0f64; for index in 0..unique_label.len() { let current_label = unique_label[index]; let current_confidence = occurrence[index].len() as f64 / sum; if current_confidence > most_confidence { most_confidence = current_confidence; current_outcome.label = current_label; } current_outcome.detail.push(OutcomeDetail { label: current_label, confidence: current_confidence }) } has_left_boundary = true; left_boundary = current_split; if s == splits.len() - 1 { current_outcome.left_boundary = Some(current_split); current_outcome.right_boundary = None; } outcome.push(current_outcome); } } outcome } /// select a sub vector from vector with given index pub trait VecMaskIndex<Target = Self> { fn select(&self, with_index: &[usize]) -> Target; } impl <T: Clone> VecMaskIndex<Vec<T>> for Vec<T> { fn select(&self, with_index: &[usize]) -> Vec<T> { let mut ret = Vec::new(); for index in with_index { ret.push(self[*index].clone()); } ret } } /// compute variance for vector with numerical data (or any data type can `Into` f64) pub trait Variance<Target = Self> { fn var(&self) -> f64; } impl <T: Clone + Into<f64>> Variance<Vec<T>> for Vec<T> { fn var(&self) -> f64 { if self.len() <= 1 { 0f64 } else { let mut avg = 0f64; for ele in self { avg += (*ele).clone().into(); } avg /= self.len() as f64; let mut sum = 0f64; for ele in self { sum += f64::powf((*ele).clone().into() - avg, 2.0f64); } sum / (self.len() as f64 - 1.0f64) } } } /// get the index of the minimal element in vector pub trait Argmin<Target = Self> { fn argmin(&self) -> Option<usize>; } impl <T: PartialOrd + Num + Clone> Argmin<Vec<T>> for Vec<T> { fn argmin(&self) -> Option<usize> { if self.len() == 0 { None } else { let mut current_min = self[0].clone(); let mut current_min_index = 0usize; for i in 0..self.len() { if current_min > self[i] { current_min = self[i].clone(); current_min_index = i; } } Some(current_min_index) } } }